Projects
|
The goal of MSB is to provide expertise on state-of-the-art methods for the analysis of the most important high-throughput experiments and for mathematical modelling for cooperation partners. We also develop customized methods that go beyond standard analyses and pipelines. In doing so, we try to transfer findings and methodological approaches for data analysis to new fields of research. |
Analysis Pipelines for High-Throughput Data
In this project, we apply and optimize comprehensive data analysis pipelines from raw data to high-level statistical analyses. We generate realistically simulated data to select the optimal analysis strategy. We also investigate the sensitivity/robustness of results to choice of algorithms and configuration parameters.
Responsible: Eva Brombacher, Eva Kohnert, Carlotta Meyring, Clemens Kreutz
Collaborators: Oliver Schilling, Fabian Cieplik
Statistical and bioinformatic analyses of proteomics and other omics data
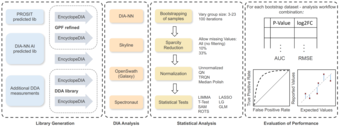
In this project, we develop computational approaches for analysis of mass-spectrometry based proteomics data. We also benchmark approaches, in particular methods for data-independent acquisition (DIA).
Responsible: Eva Brombacher, Clemens Kreutz
Our collaborators: Oliver Schilling
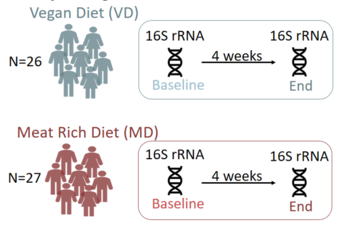
Statistical and bioinformatic analyses of microbiome data
In this project, we develop and apply statistical approaches for analysis of sequencing-based microbiome data. We currently focus on statistical models that account for the compositional nature, excessive zero counts, overdispersion, and patient-specific effects.
Responsible: Eva Kohnert, Carlotta Meyring, Clemens Kreutz
Our collaborators: Ann-Kathrin Lederer, Fabian Cieplik, Ali Al Ahmad
Analysis of Single Cell Data
In this project, we compare the performance of statistical models and tests as well as machine learning methods (e.g. clustering) for processing of single cell data, e.g. from scRNA-seq experiments. We also establish pipelines which are robust against the choice of configuration parameters.
Responsible: Eva Kohnert, Carlotta Meyring, Clemens Kreutz
Our collaborators: Florens Lohrmann, Philipp Henneke
AI & Foundation Models for Single Cell Data
In this project, we compare the performance of AI-based models and tests (e.g. scGPT) for analyzing of single cell data, e.g. from scRNA-seq. We also fine-tune foundation models for downstream tasks such as cell type annotation, predicting the expression of unobserved genes or for unobserved biological conditions.
Responsible: Jonatan Menger, Eva Brombacher, Clemens Kreutz
Our collaborators: Andreas Raue, Wolfgang Schamel, Thomas Ott, Thomas Brox
Modelling Methods in Systems Biology
In this project, we develop and extend existing methodology for establishing mathematical models of biochemical processes in living cells. We focus on
- Development of reliable and robust optimization methods for parameter estimation
- Deriving low dimensional representations of the dynamics of regulation networks
- Further advancing the RTF modelling approach as an novel modelling approach
- Approximating differential equation models by the RTF for multi-scale modelling
- Evaluation of deep learning approaches in the context of ODE modelling
- Implemention of novel methods as R package or into the Data2Dynamics modelling framework
Responsible: Niklas Neubrand, Timo Rachel, Tim Litwin, Clemens Kreutz
Our collaborator: Olaf Groß, Jens Timmer, Jan Hasenauer
Benchmarking in Systems Biology
In this project, we evalute the performance of existing computational methods for mathematical modelling by
- Comparison of the performance of optimization methods which are applied for model calibration/parameter estimation
- Establishment of benchmark problems which can be used to assess modelling methods
- Development and extension of methodology for performing reliable benchmark studies
- Establishment of approaches for the realistic generation of simulation data
Link: Benchmark models on github
Responsible: Lukas Refisch, Janine Egert, Clemens Kreutz
Our collaborators: Jens Timmer, Jan Hausenauer
Modelling in Developmental Biology
In this project, develop spatio-temporal models of cellular signalling and cell type specification of Xenopus (cilia and mucociliary development).
Responsible: Tim Litwin, Clemens Kreutz
Collaborators: Peter Walentek
Data2Dynamics
In this project, we develop and extend the Data2Dynamics modelling environment which is a high-performance expert implementation for mathematical modelling.
Link: D2D repository at github
Responsible: Tim Litwin, Niklas Neubrand, Timo Rachel, Clemens Kreutz
Our collaborators: Jens Timmer, Andreas Raue
Analysis Pipelines for High-Throughput Data
In this project, we apply and optimize comprehensive data analysis pipelines from raw data to high-level statistical analyses. We generate realistically simulated data to select the optimal analysis strategy. We also investigate the sensitivity/robustness of results to choice of algorithms and configuration parameters.
Responsible: Eva Brombacher, Eva Kohnert, Carlotta Meyring, Clemens Kreutz
Collaborators: Oliver Schilling, Fabian Cieplik
Statistical and bioinformatic analyses of proteomics and other omics data
In this project, we develop computational approaches for analysis of mass-spectrometry based proteomics data. We also benchmark approaches, in particular methods for data-independent acquisition (DIA).
Responsible: Eva Brombacher, Clemens Kreutz
Our collaborators: Oliver Schilling
Statistical and bioinformatic analyses of microbiome data
In this project, we develop and apply statistical approaches for analysis of sequencing-based microbiome data. We currently focus on statistical models that account for the compositional nature, excessive zero counts, overdispersion, and patient-specific effects.
Responsible: Eva Kohnert, Carlotta Meyring, Clemens Kreutz
Our collaborators: Ann-Kathrin Lederer, Fabian Cieplik, Ali Al Ahmad
Analysis of Single Cell Data
In this project, we compare the performance of statistical models and tests as well as machine learning methods (e.g. clustering) for processing of single cell data, e.g. from scRNA-seq experiments. We also establish pipelines which are robust against the choice of configuration parameters.
Responsible: Eva Kohnert, Carlotta Meyring, Clemens Kreutz
Our collaborators: Florens Lohrmann, Philipp Henneke
AI & Foundation Models for Single Cell Data
In this project, we compare the performance of AI-based models and tests (e.g. scGPT) for analyzing of single cell data, e.g. from scRNA-seq. We also fine-tune foundation models for downstream tasks such as cell type annotation, predicting the expression of unobserved genes or for unobserved biological conditions.
Responsible: Jonatan Menger, Eva Brombacher, Clemens Kreutz
Our collaborators: Andreas Raue, Wolfgang Schamel, Thomas Ott, Thomas Brox
Modelling Methods in Systems Biology
In this project, we develop and extend existing methodology for establishing mathematical models of biochemical processes in living cells. We focus on
- Development of reliable and robust optimization methods for parameter estimation
- Deriving low dimensional representations of the dynamics of regulation networks
- Further advancing the RTF modelling approach as an novel modelling approach
- Approximating differential equation models by the RTF for multi-scale modelling
- Evaluation of deep learning approaches in the context of ODE modelling
- Implemention of novel methods as R package or into the Data2Dynamics modelling framework
Responsible: Niklas Neubrand, Timo Rachel, Tim Litwin, Clemens Kreutz
Our collaborator: Olaf Groß, Jens Timmer, Jan Hasenauer
Benchmarking in Systems Biology
In this project, we evalute the performance of existing computational methods for mathematical modelling by
- Comparison of the performance of optimization methods which are applied for model calibration/parameter estimation
- Establishment of benchmark problems which can be used to assess modelling methods
- Development and extension of methodology for performing reliable benchmark studies
- Establishment of approaches for the realistic generation of simulation data
Link: Benchmark models on github
Responsible: Lukas Refisch, Janine Egert, Clemens Kreutz
Our collaborators: Jens Timmer, Jan Hausenauer
Modelling in Developmental Biology
In this project, develop spatio-temporal models of cellular signalling and cell type specification of Xenopus (cilia and mucociliary development).
Responsible: Tim Litwin, Clemens Kreutz
Collaborators: Peter Walentek
Data2Dynamics
In this project, we develop and extend the Data2Dynamics modelling environment which is a high-performance expert implementation for mathematical modelling.
Link: D2D repository at github
Responsible: Tim Litwin, Niklas Neubrand, Timo Rachel, Clemens Kreutz
Our collaborators: Jens Timmer, Andreas Raue
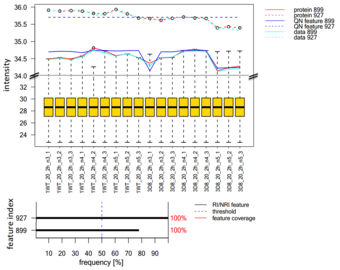
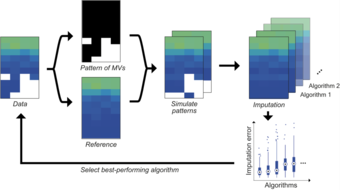
Imputation and normalization for proteomics data
In this project, we focus on dealing optimally with missing values and on data preprocessing like normalization.
Responsible: Eva Brombacher, Janine Egert, Clemens Kreutz
Our collaborators: Oliver Schilling, Bettina Warscheid
Analysis of Mass Cytometry Data
In this project, we compare the performance of statistical models and tests as well as machine learning methods for processing of mass cytometry data.
Responsible: Ariane Schad, Clemens Kreutz
Our collaborators: Stefan Reinker (Novartis)
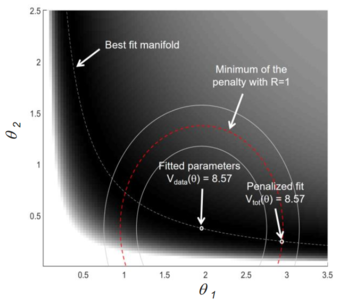
Modelling for assessing parameter uncertainties
In this project, we derive a valid statistical methodology for parameter and prediction uncertainties as well as for identifiablity and observability analyses. Furthermore, we establish strategies like L1 regularization for deriving small minimal models, such as ITRP.
Responsible: Lukas Refisch, Rafael Arutjunjan, Clemens Kreutz
Our collaborator: Jens Timmer, Andreas Raue
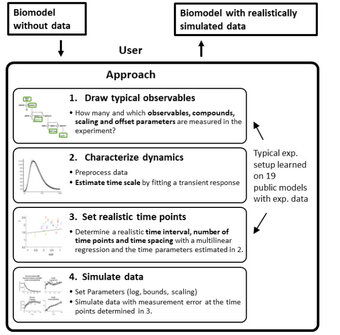
Benchmarking problems in Systems Biology and realistically simulated data
In this project, we establish benchmark problems which can be used to assess modelling methods and approaches for the realistic generation of simulation data.
Link: Benchmark models on github
Responsible: Lukas Refisch, Janine Egert, Clemens Kreutz
Our collaborators: Jens Timmer, Jan Hausenauer
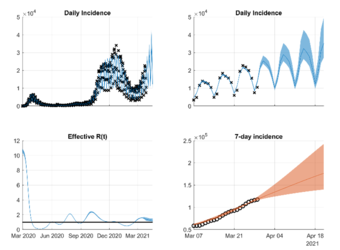
Prediction of local COVID-19 progression
In this project, we analyze the IfSG data of the confirmed COVID-19 cases at the level of counties (Landkreise) and provide daily updated predictions for new infections for all ICUs in Germany.
Responsible: Lukas Refisch, Fabian Lorenz, Clemens Kreutz
Our collaborators: Robert Koch Institute (RKI), Federal Institute for Population Research (BIB), German Aerospace Center (DLR)
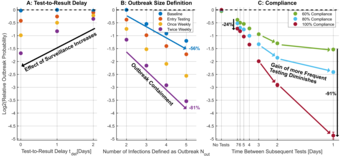
COVID-19 Surveillance of Hospitalized Patients
In this project, we optimize surveillance and test strategies for patients and employees in the Oberbergkliniken based on agent-based stochastical dynamical models.
Responsible: Tim Litwin, Jens Timmer, Clemens Kreutz
Our collaborators: Matthias Müller, Andreas Wahl Kordon, Marcus Panning, Mathias Berger
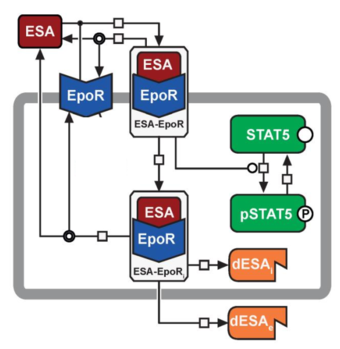
Optimal Personalized Treatment of Anemia
In this project, we apply a mathematical model of EPO binding and its effect on erythropoesis to predict the optimal time- and dosage strategy for treating anemia. We consider cancer as well as patients with chronic kidney disease.
Responsible: Lukas Refisch, Clemens Kreutz
Our collaborators: Ursula Kingmüller, Jens Timmer
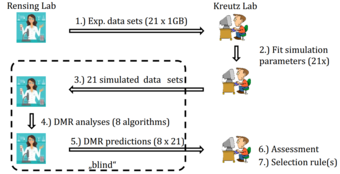
Identification of Differentially Methylated Regions (DMRs) from Bisulfite Sequencing (BSSEQ) Data
In this project, we compare and assess the performances of algorithms for detecting differentially methylated regions (DMRs) from bisulfite sequencing (BSSEQ) data.
Responsible: Clemens Kreutz
Our collaborators: Stefan Rensing
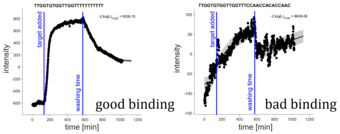
Optimizing adaptamer arrays
In this project, we analyze kinetic data of adaptamer binding and dissociation by mathematical models. We assess binding characteristics and develop predictive models for testing new adaptamer sequences.
Responsible: Lukas Refisch, Clemens Kreutz
Our collaborator: Günter Roth (BioCopy)
Benchmarking Wiki
In this project, we started to develop a wiki as a repository for published benchmark studies. We intend to collect insights from literature obtained by comparing the performance of computational approaches.
The benchmarking wiki also offers the opportunity to publish (possibly small or unpublished) own results.
Everybody is welcome to contribute!
Link: Benchmark Wiki
Responsible: Clemens Kreutz
Imputation and normalization for proteomics data
In this project, we focus on dealing optimally with missing values and on data preprocessing like normalization.
Responsible: Eva Brombacher, Janine Egert, Clemens Kreutz
Our collaborators: Oliver Schilling, Bettina Warscheid
Analysis of Mass Cytometry Data
In this project, we compare the performance of statistical models and tests as well as machine learning methods for processing of mass cytometry data.
Responsible: Ariane Schad, Clemens Kreutz
Our collaborators: Stefan Reinker (Novartis)
Modelling for assessing parameter uncertainties
In this project, we derive a valid statistical methodology for parameter and prediction uncertainties as well as for identifiablity and observability analyses. Furthermore, we establish strategies like L1 regularization for deriving small minimal models, such as ITRP.
Responsible: Lukas Refisch, Rafael Arutjunjan, Clemens Kreutz
Our collaborator: Jens Timmer, Andreas Raue
Benchmarking problems in Systems Biology and realistically simulated data
In this project, we establish benchmark problems which can be used to assess modelling methods and approaches for the realistic generation of simulation data.
Link: Benchmark models on github
Responsible: Lukas Refisch, Janine Egert, Clemens Kreutz
Our collaborators: Jens Timmer, Jan Hausenauer
Prediction of local COVID-19 progression
In this project, we analyze the IfSG data of the confirmed COVID-19 cases at the level of counties (Landkreise) and provide daily updated predictions for new infections for all ICUs in Germany.
Responsible: Lukas Refisch, Fabian Lorenz, Clemens Kreutz
Our collaborators: Robert Koch Institute (RKI), Federal Institute for Population Research (BIB), German Aerospace Center (DLR)
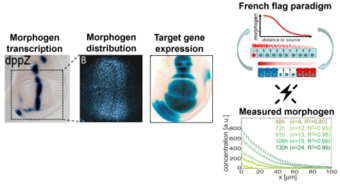
Modelling of Pattern Formation
In this project, develop spatio-temporal models of cellular signalling during morphogenesis in Drosophila (wing formation) and Xenopus (cilia and mucociliary development).
Responsible: Fabian Lorenz, Rafael Arutjunjan, Janine Egert, Clemens Kreutz
Collaborators: Georgios Pyrowolakis, Peter Walentek
COVID-19 Surveillance of Hospitalized Patients
In this project, we optimize surveillance and test strategies for patients and employees in the Oberbergkliniken based on agent-based stochastical dynamical models.
Responsible: Tim Litwin, Jens Timmer, Clemens Kreutz
Our collaborators: Matthias Müller, Andreas Wahl Kordon, Marcus Panning, Mathias Berger
Optimal Personalized Treatment of Anemia
In this project, we apply a mathematical model of EPO binding and its effect on erythropoesis to predict the optimal time- and dosage strategy for treating anemia. We consider cancer as well as patients with chronic kidney disease.
Responsible: Lukas Refisch, Clemens Kreutz
Our collaborators: Ursula Kingmüller, Jens Timmer
Identification of Differentially Methylated Regions (DMRs) from Bisulfite Sequencing (BSSEQ) Data
In this project, we compare and assess the performances of algorithms for detecting differentially methylated regions (DMRs) from bisulfite sequencing (BSSEQ) data.
Responsible: Clemens Kreutz
Our collaborators: Stefan Rensing
Optimizing adaptamer arrays
In this project, we analyze kinetic data of adaptamer binding and dissociation by mathematical models. We assess binding characteristics and develop predictive models for testing new adaptamer sequences.
Responsible: Lukas Refisch, Clemens Kreutz
Our collaborator: Günter Roth (BioCopy)
Benchmarking Wiki
In this project, we started to develop a wiki as a repository for published benchmark studies. We intend to collect insights from literature obtained by comparing the performance of computational approaches.
The benchmarking wiki also offers the opportunity to publish (possibly small or unpublished) own results.
Everybody is welcome to contribute!
Link: Benchmark Wiki
Responsible: Clemens Kreutz