Medical Epigenetics - Arbeitsgruppe Timmers
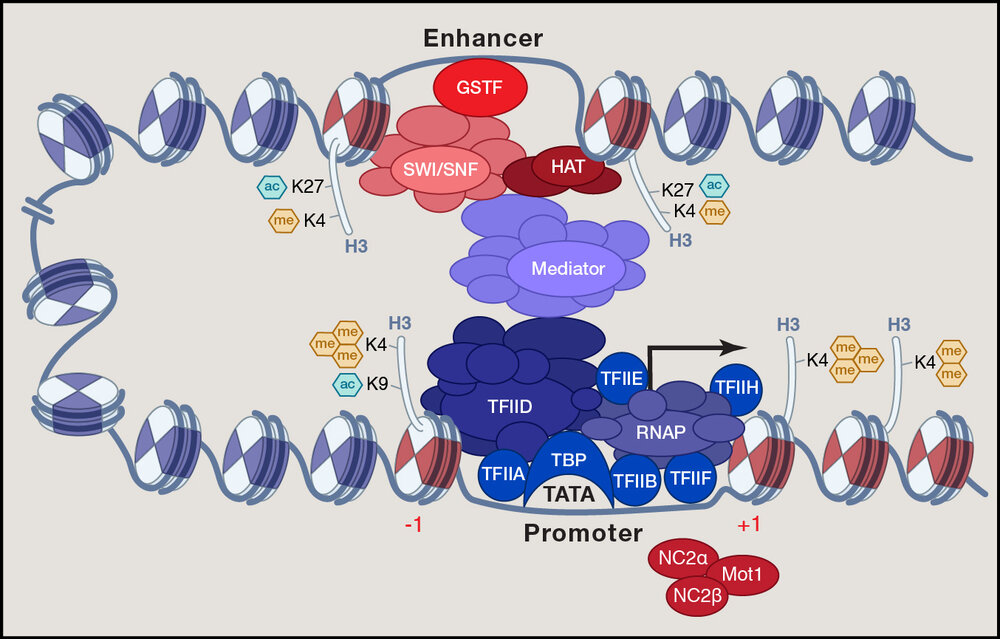
Prof. Dr. Marc TimmersModel of the three different levels of transcription initiation control and of associated chromatin modifications (taken from Koster et al. 2015 Cell 161:724).
Cancer-genome sequencing projects led to the realization that epigenetic pathways are often disturbed by gene mutations in solid cancers. The epigenetic control is exerted at the level of the structure and modification of chromatin and disturbances directly impacts tissue-specific programs of gene transcription and, hence, of cell identity and behaviour. An attractive aspect of epigenetic control for effective intervention strategies in solid cancers is the dynamic equilibrium between epigenetic states and the specificity and selectivity of the enzymes determining this equilibrium.
Our group studies transcriptional regulation in relation to chromatin modifications which are relevant to human diseases. In particular, we focus on the MLL complexes which deposit histone methylation and regulate the activity of gene enhancers and gene promoters. Genes encoding integral subunits of the MLL3 and MLL4 complexes display the highest mutational frequency in bladder cancer. This type of tumor is characterized by a strong gender imbalance and we are examining the mechanistic basis for this. Besides these molecular studies, we are performing both genetic and chemical screens to determine targetable vulnerabilities of bladder cancer cell lines and primary 2D and 3D cultures from bladder cancers with the aim to develop new treatment options for this disease. The menin subunit of the MLL1/MLL2 complexes has become a major focus for leukemia research as chemical menin-MLL inhibitors display remarkable efficacy in certain types of AML and are expected to receive FDA approval for treatment soon. All cellular and epigenetic signaling culminates in the loading of RNA polymerase II to specific sets of gene promoters. The basal transcription factor TFIID plays a coordinating role in setting transcription initiation frequencies. We are examining the chromatin aspects on the dynamic regulation of TFIID activity.
In general, our studies of gene transcription and chromatin regulation employ combinations of cell biology, molecular biology, proteomics, genomics and bioinformatics approaches.
Future projects and goals
- Regulation and function of histone H3K4 methylation in gene transcription programs
- Understanding the impact of oncogenic lesions on histone methylation pathways
- Development and application of drugs targeting epigenetic pathways disturbed in solid tumors
FREpi Library

Interested in contributing to our research?
Motivated and ambitious researchers at all levels (M.Sc. and Ph.D. students, post-doctoral fellows) are encouraged to contact us (m.timmers -at- dkfz-heidelberg.de or baerbel.hansen -at- dkfz-heidelberg.de) for open positions.